Joining Datasets
Joining is needed when you have two or more corresponding datasets, each dataset having some parts to compose each sample.
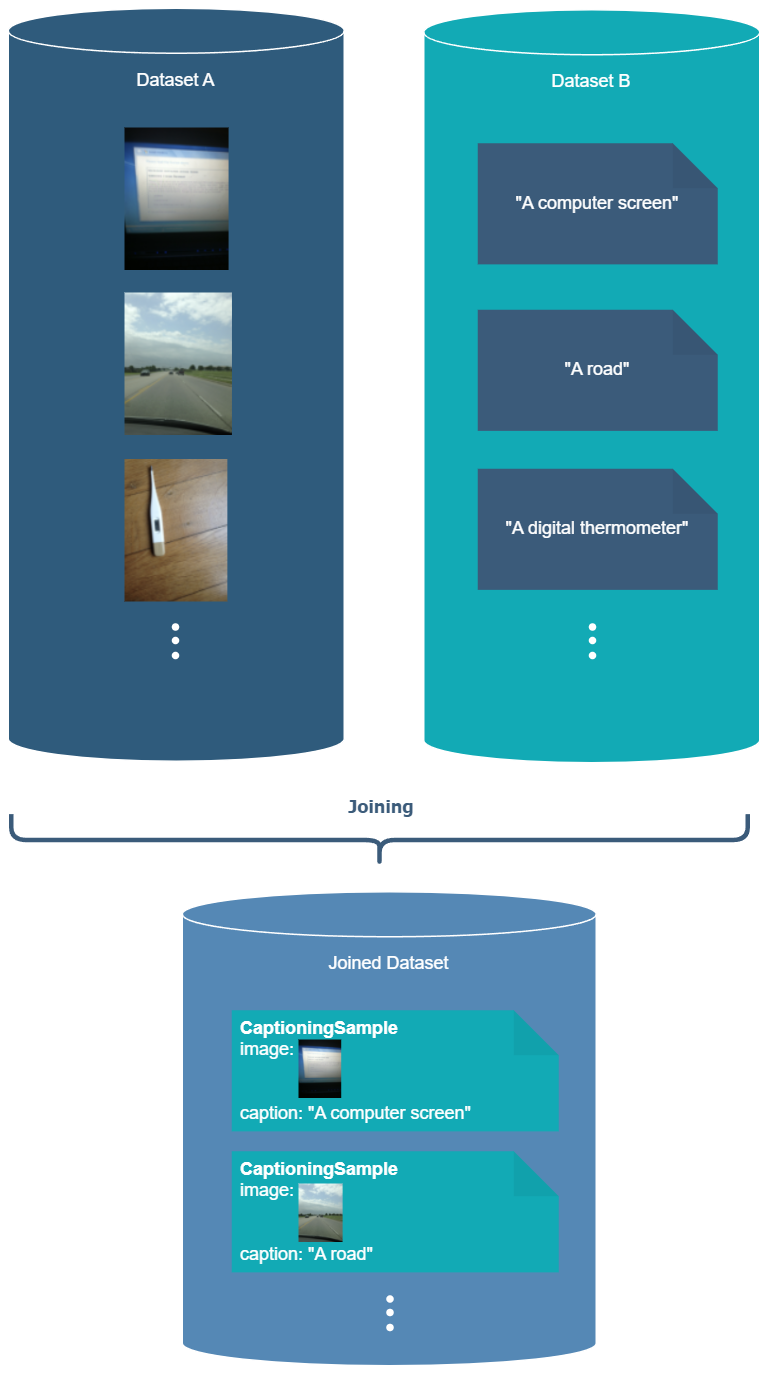
Dataset joining is analogous to webdataset’s “add column”, i.e., merging two datasets which require the same files within the tar files.
Example File Structure
Here is an example structure of the datasets within the tar files:
ds1
├── .nv-meta
│ ├── .info.json
│ ├── split.yaml
│ └── dataset.yaml
├── shard1.tar
│ ├── 0001.jpg
│ ├── 0002.jpg
│ └── 0003.jpg
├── shard1.idx
ds2
├── .nv-meta
│ ├── .info.json
│ ├── split.yaml
│ └── dataset.yaml
├── shard1.tar
│ ├── 0001.txt
│ ├── 0002.txt
│ └── 0003.txt
├── shard1.idx
In this example, ds1/shard1.tar and ds2/shard1.tar contain files with the same base names / keys.
When joining, the files from both datasets are joined based on their keys, which must be unique within each dataset across all the shards. Each dataset must be prepared (i.e. .nv-meta created) with a recent version of energon.
Minimal Example
Below is a minimal meta dataset example for joining.
metadataset_minimal.yaml:
__module__: megatron.energon
__class__: MetadatasetV2
splits:
train:
join:
- path: ds1
- path: ds2
joiner:
__module__: megatron.energon
__class__: CaptioningSample
Note that the blending is omitted here, as it is optional. Joining is of course also optional.
Warning
All metadatasets that contain a join must be prepared with the energon prepare command.
This will compute the join index and store it next to the metadataset in a folder with a similar name.
Join Modes
When joining two datasets, it may happen that the first dataset (primary dataset) has more samples or fewer samples than the secondary dataset(s). In this case, we have to decide how to handle these samples that do not match.
The primary dataset always serves as the reference and there will never be more samples in the join result than in the primary dataset. However if a primary sample has no match in a secondary dataset, it may be skipped as explained below.
For each of the secondary datasets, the user can specify a nonmatch setting.
With one of the following options, the user can decide what happens, if a sample from the primary dataset is not found in the given secondary dataset:
error(default): An error is raisedskip: The whole sample is skippednone: The column for the current secondary dataset is filled withNoneif there’s no match
Example metadataset_nomatch.yaml:
__module__: megatron.energon
__class__: MetadatasetV2
splits:
train:
join:
- path: ds1
- path: ds2
nonmatch: skip
- path: ds3
nonmatch: none
joiner:
__module__: megatron.energon
__class__: CaptioningSample
To illustrate the effect, let’s look at some example data:
ds1samples:s1,s2,s3,s5,s6ds2samples:s1,s3,s4,s6,s7ds3samples:s1,s2,s3,s100
The resulting joined data would contain the following samples, one in each row:
ds1 |
ds2 |
ds3 |
|---|---|---|
s1 |
s1 |
s1 |
s3 |
s3 |
s3 |
s6 |
s6 |
None |
Explanation:
The sample key
s1is available in all dataset.s2is missing fromds2and nonmatch is set toskip, so the sample will not appear in the result.s3is available in all datasets.s4is not in the primary dataset. Only samples from the primary dataset will be included.s5is missing fromds2again, and this time also fromds3s6is missing fromds3andds3hasnonmatchset tonone, so the sample is not skipped, but the column fords3is set toNone
Extensive Example
Here is a more extensive example that shows multiple things at once:
Joining can be used inside blending
The datasets to be joined can have custom subflavors or dataset yamls specified
A custom “joiner” can be specified to define how samples are joined and what the resulting type is
The
nonmatchsetting is not included here, but would work just like shown above
metadataset_extended.yaml:
__module__: megatron.energon
__class__: MetadatasetV2
splits:
train:
blend:
- weight: 1
join:
- path: ds1
dataset_config: dataset.yaml # If override is needed
- path: ds2
dataset_config: dataset.yaml
subflavors: # If needed, will be merged(overriding) with parent subflavor
ds2_extra: 2
split_config: split.yaml
joiner:
__module__: my_module
__class__: JoinedSample # Type should implement from_joined(ds1, ds2)
split_config: split.yaml # Sets this for all joined datasets
split_part: train # Sets this for all joined datasets
subflavors: # Sets this for all joined datasets (it will be merged with their individual subflavors)
source: metadataset.yaml
src: ds1
Custom Join Type
To define a custom join type, you can create a Python class as shown below in my_module.py:
from dataclasses import dataclass
import torch
from megatron.energon import Sample, TextSample
@dataclass
class JoinedSample(Sample):
text1: torch.Tensor
text2: torch.Tensor
@staticmethod
def from_joined(ds1: TextSample, ds2: TextSample) -> "JoinedSample":
return JoinedSample.derive_from(
ds1,
text1=ds1.text,
text2=ds2.text,
)
This class should implement the from_joined method to combine samples from ds1 and ds2.
Note: It is important to use derive_from with the first argument being the first sample, as this will guarantee that the state can be saved and restored. It ensures that all the internal keys of the sample are retained.