Note
Go to the end to download the full example code
Extending Diagnostic Models#
Implementing a custom diagnostic model
This example will demonstrate how to extend Earth2Studio by implementing a custom diagnostic model and running it in a general workflow.
In this example you will learn:
API requirements of diagnostic models
Implementing a custom diagnostic model
Running this custom model in a workflow with built in prognostic
Custom Diagnostic#
As discussed in the Diagnostic Models section of the user guide,
Earth2Studio defines a diagnostic model through a simple interface
earth2studio.models.dx.base.Diagnostic Model. This can be used to help
guide the required APIs needed to successfully create our own model.
In this example, lets consider a simple diagnostic that converts the surface temperature in Kelvin to Celsius to make it more readable for the average person.
Our diagnostic model has a base class of torch.nn.Module which allows us
to get the required to(device) method for free.
from collections import OrderedDict
import numpy as np
import torch
from earth2studio.models.batch import batch_func
from earth2studio.utils import handshake_coords, handshake_dim
from earth2studio.utils.type import CoordSystem
class CustomDiagnostic(torch.nn.Module):
"""Custom dianostic model"""
def __init__(self):
super().__init__()
input_coords = OrderedDict(
{
"batch": np.empty(1),
"variable": np.array(["t2m"]),
"lat": np.linspace(90, -90, 721),
"lon": np.linspace(0, 360, 1440, endpoint=False),
}
)
output_coords = OrderedDict(
{
"batch": np.empty(1),
"variable": np.array(["t2m_c"]),
"lat": np.linspace(90, -90, 721),
"lon": np.linspace(0, 360, 1440, endpoint=False),
}
)
@batch_func()
def __call__(
self,
x: torch.Tensor,
coords: CoordSystem,
) -> tuple[torch.Tensor, CoordSystem]:
"""Runs diagnostic model
Parameters
----------
x : torch.Tensor
Input tensor
coords : CoordSystem
Input coordinate system
"""
for i, (key, value) in enumerate(self.input_coords.items()):
if key != "batch":
handshake_dim(coords, key, i)
handshake_coords(coords, self.input_coords, key)
out_coords = coords.copy()
out_coords["variable"] = self.output_coords["variable"]
out = x - 273.15 # To celcius
return out, out_coords
Input/Output Coordinates#
Defining the input/output coordinate systems is essential for any model in Earth2Studio since this is how both the package and users can learn what type of data the model expects. Have a look at Coordinate Systems for details on coordinate system. For this diagnostic model, we simply define the input coordinates to be the global surface temperature specified in :py:file:`earth2studio.lexicon.base.py`. The output is a custom variable :py:var:`t2m_c` that represents the temperature in Celsius.
__call__() API#
The call function is the main API of diagnostic models that have a tensor and coordinate system as input/output. This function first validates that the coordinate system is correct. Then both the input data tensor and also coordinate system are updated and returned.
Note
You may notice the batch_func() decorator, which is used to make batched
operations easier. For more details about this refer to the batch_function_userguide
section of the user guide.
Set Up#
With the custom diagnostic model defined, it’s now easily usable in a workflow. Let’s create our own simple diagnostic workflow based on the ones that exist already in Earth2Studio.
from datetime import datetime
from typing import Optional
import numpy as np
import torch
from loguru import logger
from tqdm import tqdm
from earth2studio.data import DataSource, fetch_data
from earth2studio.io import IOBackend
from earth2studio.models.dx import DiagnosticModel
from earth2studio.models.px import PrognosticModel
from earth2studio.utils.coords import extract_coords, map_coords
from earth2studio.utils.time import to_time_array
def run(
time: list[str] | list[datetime] | list[np.datetime64],
nsteps: int,
prognostic: PrognosticModel,
diagnostic: DiagnosticModel,
data: DataSource,
io: IOBackend,
device: Optional[torch.device] = None,
) -> IOBackend:
"""Simple diagnostic workflow
Parameters
----------
time : list[str] | list[datetime] | list[np.datetime64]
List of string, datetimes or np.datetime64
nsteps : int
Number of forecast steps
prognostic : PrognosticModel
Prognostic models
data : DataSource
Data source
io : IOBackend
IO object
device : Optional[torch.device], optional
Device to run inference on, by default None
Returns
-------
IOBackend
Output IO object
"""
logger.info("Running diagnostic workflow!")
# Load model onto the device
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
logger.info(f"Inference device: {device}")
prognostic = prognostic.to(device)
# Fetch data from data source and load onto device
time = to_time_array(time)
x, coords = fetch_data(
source=data,
time=time,
lead_time=prognostic.input_coords["lead_time"],
variable=prognostic.input_coords["variable"],
device=device,
)
logger.success(f"Fetched data from {data.__class__.__name__}")
# Set up IO backend
total_coords = prognostic.output_coords.copy()
del total_coords["batch"] # Unsafe if batch not supported
for key, value in total_coords.items():
if value.shape == 0:
del total_coords[key]
total_coords["time"] = time
total_coords["lead_time"] = np.asarray(
[prognostic.output_coords["lead_time"] * i for i in range(nsteps + 1)]
).flatten()
total_coords.move_to_end("lead_time", last=False)
total_coords.move_to_end("time", last=False)
for name, value in diagnostic.output_coords.items():
if name == "batch":
continue
total_coords[name] = value
var_names = total_coords.pop("variable")
io.add_array(total_coords, var_names)
# Map lat and lon if needed
x, coords = map_coords(x, coords, prognostic.input_coords)
# Create prognostic iterator
model = prognostic.create_iterator(x, coords)
logger.info("Inference starting!")
with tqdm(total=nsteps + 1, desc="Running inference") as pbar:
for step, (x, coords) in enumerate(model):
# Run diagnostic
x, coords = map_coords(x, coords, diagnostic.input_coords)
x, coords = diagnostic(x, coords)
io.write(*extract_coords(x, coords))
pbar.update(1)
if step == nsteps:
break
logger.success("Inference complete")
return io
Lets instantiate the components needed.
Prognostic Model: Use the built in DLWP model
earth2studio.models.px.DLWP.Diagnostic Model: The custom diagnostic model defined above
Datasource: Pull data from the GFS data api
earth2studio.data.GFS.IO Backend: Save the outputs into a Zarr store
earth2studio.io.ZarrBackend.
from collections import OrderedDict
import numpy as np
from dotenv import load_dotenv
load_dotenv() # TODO: make common example prep function
from earth2studio.data import GFS
from earth2studio.io import ZarrBackend
from earth2studio.models.px import DLWP
# Load the default model package which downloads the check point from NGC
package = DLWP.load_default_package()
model = DLWP.load_model(package)
# Diagnostic model
diagnostic = CustomDiagnostic()
# Create the data source
data = GFS()
# Create the IO handler, store in memory
io = ZarrBackend()
Execute the Workflow#
Running our workflow with a build in prognostic model and a custom diagnostic is as simple as the following.
nsteps = 20
io = run(["2024-01-01"], nsteps, model, diagnostic, data, io)
print(io.root.tree())
2024-04-19 00:36:48.079 | INFO | __main__:run:189 - Running diagnostic workflow!
2024-04-19 00:36:48.079 | INFO | __main__:run:192 - Inference device: cuda
2024-04-19 00:36:48.086 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:151 - Fetching GFS index file: 2023-12-31 18:00:00
Fetching GFS for 2023-12-31 18:00:00: 0%| | 0/7 [00:00<?, ?it/s]
2024-04-19 00:36:48.188 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: t850 at 2023-12-31 18:00:00
Fetching GFS for 2023-12-31 18:00:00: 0%| | 0/7 [00:00<?, ?it/s]
2024-04-19 00:36:48.208 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: z1000 at 2023-12-31 18:00:00
Fetching GFS for 2023-12-31 18:00:00: 0%| | 0/7 [00:00<?, ?it/s]
2024-04-19 00:36:48.227 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: z700 at 2023-12-31 18:00:00
Fetching GFS for 2023-12-31 18:00:00: 0%| | 0/7 [00:00<?, ?it/s]
2024-04-19 00:36:48.250 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: z500 at 2023-12-31 18:00:00
Fetching GFS for 2023-12-31 18:00:00: 0%| | 0/7 [00:00<?, ?it/s]
2024-04-19 00:36:48.270 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: z300 at 2023-12-31 18:00:00
Fetching GFS for 2023-12-31 18:00:00: 0%| | 0/7 [00:00<?, ?it/s]
Fetching GFS for 2023-12-31 18:00:00: 71%|███████▏ | 5/7 [00:00<00:00, 49.90it/s]
2024-04-19 00:36:48.288 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: tcwv at 2023-12-31 18:00:00
Fetching GFS for 2023-12-31 18:00:00: 71%|███████▏ | 5/7 [00:00<00:00, 49.90it/s]
2024-04-19 00:36:48.307 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: t2m at 2023-12-31 18:00:00
Fetching GFS for 2023-12-31 18:00:00: 71%|███████▏ | 5/7 [00:00<00:00, 49.90it/s]
Fetching GFS for 2023-12-31 18:00:00: 100%|██████████| 7/7 [00:00<00:00, 50.96it/s]
2024-04-19 00:36:48.336 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:151 - Fetching GFS index file: 2024-01-01 00:00:00
Fetching GFS for 2024-01-01 00:00:00: 0%| | 0/7 [00:00<?, ?it/s]
2024-04-19 00:36:48.422 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: t850 at 2024-01-01 00:00:00
Fetching GFS for 2024-01-01 00:00:00: 0%| | 0/7 [00:00<?, ?it/s]
2024-04-19 00:36:48.443 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: z1000 at 2024-01-01 00:00:00
Fetching GFS for 2024-01-01 00:00:00: 0%| | 0/7 [00:00<?, ?it/s]
2024-04-19 00:36:48.464 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: z700 at 2024-01-01 00:00:00
Fetching GFS for 2024-01-01 00:00:00: 0%| | 0/7 [00:00<?, ?it/s]
2024-04-19 00:36:48.483 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: z500 at 2024-01-01 00:00:00
Fetching GFS for 2024-01-01 00:00:00: 0%| | 0/7 [00:00<?, ?it/s]
2024-04-19 00:36:48.502 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: z300 at 2024-01-01 00:00:00
Fetching GFS for 2024-01-01 00:00:00: 0%| | 0/7 [00:00<?, ?it/s]
2024-04-19 00:36:48.520 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: tcwv at 2024-01-01 00:00:00
Fetching GFS for 2024-01-01 00:00:00: 0%| | 0/7 [00:00<?, ?it/s]
Fetching GFS for 2024-01-01 00:00:00: 86%|████████▌ | 6/7 [00:00<00:00, 50.53it/s]
2024-04-19 00:36:48.541 | DEBUG | earth2studio.data.gfs:fetch_gfs_dataarray:197 - Fetching GFS grib file for variable: t2m at 2024-01-01 00:00:00
Fetching GFS for 2024-01-01 00:00:00: 86%|████████▌ | 6/7 [00:00<00:00, 50.53it/s]
Fetching GFS for 2024-01-01 00:00:00: 100%|██████████| 7/7 [00:00<00:00, 49.68it/s]
2024-04-19 00:36:48.599 | SUCCESS | __main__:run:203 - Fetched data from GFS
2024-04-19 00:36:48.617 | INFO | __main__:run:231 - Inference starting!
Running inference: 0%| | 0/21 [00:00<?, ?it/s]
Running inference: 10%|▉ | 2/21 [00:00<00:01, 13.33it/s]
Running inference: 19%|█▉ | 4/21 [00:00<00:01, 12.94it/s]
Running inference: 29%|██▊ | 6/21 [00:00<00:01, 12.31it/s]
Running inference: 38%|███▊ | 8/21 [00:00<00:01, 12.25it/s]
Running inference: 48%|████▊ | 10/21 [00:00<00:00, 11.85it/s]
Running inference: 57%|█████▋ | 12/21 [00:00<00:00, 11.67it/s]
Running inference: 67%|██████▋ | 14/21 [00:01<00:00, 11.32it/s]
Running inference: 76%|███████▌ | 16/21 [00:01<00:00, 11.14it/s]
Running inference: 86%|████████▌ | 18/21 [00:01<00:00, 10.92it/s]
Running inference: 95%|█████████▌| 20/21 [00:01<00:00, 10.88it/s]
Running inference: 100%|██████████| 21/21 [00:01<00:00, 11.45it/s]
2024-04-19 00:36:50.452 | SUCCESS | __main__:run:244 - Inference complete
/
├── lat (721,) float64
├── lead_time (21,) timedelta64[h]
├── lon (1440,) float64
├── t2m_c (1, 21, 721, 1440) float32
└── time (1,) datetime64[ns]
Post Processing#
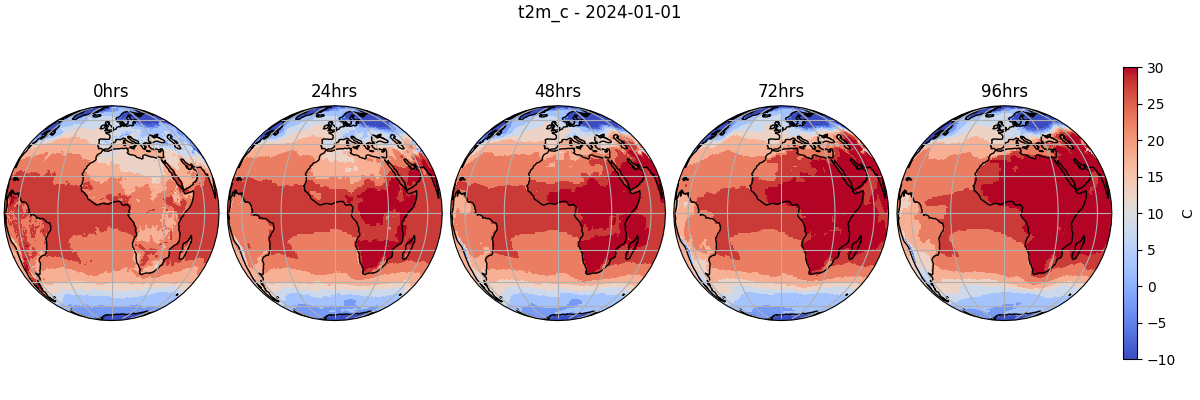
Let’s plot the Celsius temperature field from our custom diagnostic model.
import os
os.makedirs("outputs", exist_ok=True)
import cartopy.crs as ccrs
import matplotlib.pyplot as plt
forecast = "2024-01-01"
variable = "t2m_c"
plt.close("all")
# Create a figure and axes with the specified projection
fig, ax = plt.subplots(
1,
5,
figsize=(12, 4),
subplot_kw={"projection": ccrs.Orthographic()},
constrained_layout=True,
)
times = io["lead_time"].astype("timedelta64[h]").astype(int)
step = 4 # 24hrs
for i, t in enumerate(range(0, 20, step)):
ctr = ax[i].contourf(
io["lon"][:],
io["lat"][:],
io[variable][0, t],
vmin=-10,
vmax=30,
transform=ccrs.PlateCarree(),
levels=20,
cmap="coolwarm",
)
ax[i].set_title(f"{times[t]}hrs")
ax[i].coastlines()
ax[i].gridlines()
plt.suptitle(f"{variable} - {forecast}")
cbar = plt.cm.ScalarMappable(cmap="coolwarm")
cbar.set_array(io[variable][0, 0])
cbar.set_clim(-10.0, 30)
cbar = fig.colorbar(cbar, ax=ax[-1], orientation="vertical", label="C", shrink=0.8)
plt.savefig("outputs/custom_diagnostic_dlwp_prediction.jpg")
Total running time of the script: (0 minutes 43.041 seconds)
