Variational Quantum Eigensolver¶
A common application of the Variational Quantum Eigensolver (VQE) algorithm is to compute the ground state energy of a molecular system. The code below demonstrates how to perform classical preprocessing for a \(H_2\) molecule (i.e. obtain the integrals from a Hartree-Fock computation to build the molecular Hamiltonian), prepare the initial Hartree-Fock state on the quantum register, add the parameterized UCCSD ansatz to the kernel, and select the COBYLA optimizer. We are then ready to call
cudaq:vqe to estimate the minimum energy of the system.
[1]:
!pip install openfermionpyscf==0.5 matplotlib==3.8.4 scipy==1.13.0
Requirement already satisfied: openfermionpyscf==0.5 in /usr/local/lib/python3.10/dist-packages (0.5)
Requirement already satisfied: matplotlib==3.8.4 in /usr/local/lib/python3.10/dist-packages (3.8.4)
Collecting scipy==1.13.0
Downloading scipy-1.13.0-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl (38.6 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 38.6/38.6 MB 55.6 MB/s eta 0:00:00
Requirement already satisfied: openfermion>=0.5 in /usr/local/lib/python3.10/dist-packages (from openfermionpyscf==0.5) (1.6.1)
Requirement already satisfied: pytest in /usr/local/lib/python3.10/dist-packages (from openfermionpyscf==0.5) (8.2.0)
Requirement already satisfied: pyscf in /usr/local/lib/python3.10/dist-packages (from openfermionpyscf==0.5) (2.5.0)
Requirement already satisfied: contourpy>=1.0.1 in /usr/local/lib/python3.10/dist-packages (from matplotlib==3.8.4) (1.2.1)
Requirement already satisfied: packaging>=20.0 in /usr/local/lib/python3.10/dist-packages (from matplotlib==3.8.4) (24.0)
Requirement already satisfied: cycler>=0.10 in /usr/local/lib/python3.10/dist-packages (from matplotlib==3.8.4) (0.12.1)
Requirement already satisfied: python-dateutil>=2.7 in /usr/local/lib/python3.10/dist-packages (from matplotlib==3.8.4) (2.9.0.post0)
Requirement already satisfied: fonttools>=4.22.0 in /usr/local/lib/python3.10/dist-packages (from matplotlib==3.8.4) (4.51.0)
Requirement already satisfied: numpy>=1.21 in /usr/local/lib/python3.10/dist-packages (from matplotlib==3.8.4) (1.26.4)
Requirement already satisfied: pillow>=8 in /usr/local/lib/python3.10/dist-packages (from matplotlib==3.8.4) (10.3.0)
Requirement already satisfied: pyparsing>=2.3.1 in /usr/local/lib/python3.10/dist-packages (from matplotlib==3.8.4) (3.1.2)
Requirement already satisfied: kiwisolver>=1.3.1 in /usr/local/lib/python3.10/dist-packages (from matplotlib==3.8.4) (1.4.5)
Requirement already satisfied: pubchempy in /usr/local/lib/python3.10/dist-packages (from openfermion>=0.5->openfermionpyscf==0.5) (1.0.4)
Requirement already satisfied: networkx in /usr/local/lib/python3.10/dist-packages (from openfermion>=0.5->openfermionpyscf==0.5) (3.3)
Requirement already satisfied: cirq-core~=1.0 in /usr/local/lib/python3.10/dist-packages (from openfermion>=0.5->openfermionpyscf==0.5) (1.3.0)
Requirement already satisfied: requests>=2.18 in /usr/local/lib/python3.10/dist-packages (from openfermion>=0.5->openfermionpyscf==0.5) (2.31.0)
Requirement already satisfied: sympy in /usr/local/lib/python3.10/dist-packages (from openfermion>=0.5->openfermionpyscf==0.5) (1.12)
Requirement already satisfied: h5py>=2.8 in /usr/local/lib/python3.10/dist-packages (from openfermion>=0.5->openfermionpyscf==0.5) (3.10.0)
Requirement already satisfied: deprecation in /usr/local/lib/python3.10/dist-packages (from openfermion>=0.5->openfermionpyscf==0.5) (2.1.0)
Requirement already satisfied: six>=1.5 in /usr/local/lib/python3.10/dist-packages (from python-dateutil>=2.7->matplotlib==3.8.4) (1.16.0)
Requirement already satisfied: setuptools in /usr/lib/python3/dist-packages (from pyscf->openfermionpyscf==0.5) (59.6.0)
Requirement already satisfied: tomli>=1 in /usr/local/lib/python3.10/dist-packages (from pytest->openfermionpyscf==0.5) (2.0.1)
Requirement already satisfied: iniconfig in /usr/local/lib/python3.10/dist-packages (from pytest->openfermionpyscf==0.5) (2.0.0)
Requirement already satisfied: exceptiongroup>=1.0.0rc8 in /usr/local/lib/python3.10/dist-packages (from pytest->openfermionpyscf==0.5) (1.2.1)
Requirement already satisfied: pluggy<2.0,>=1.5 in /usr/local/lib/python3.10/dist-packages (from pytest->openfermionpyscf==0.5) (1.5.0)
Requirement already satisfied: tqdm in /usr/local/lib/python3.10/dist-packages (from cirq-core~=1.0->openfermion>=0.5->openfermionpyscf==0.5) (4.66.4)
Requirement already satisfied: typing-extensions>=4.2 in /usr/local/lib/python3.10/dist-packages (from cirq-core~=1.0->openfermion>=0.5->openfermionpyscf==0.5) (4.11.0)
Requirement already satisfied: pandas in /usr/local/lib/python3.10/dist-packages (from cirq-core~=1.0->openfermion>=0.5->openfermionpyscf==0.5) (2.2.2)
Requirement already satisfied: sortedcontainers~=2.0 in /usr/local/lib/python3.10/dist-packages (from cirq-core~=1.0->openfermion>=0.5->openfermionpyscf==0.5) (2.4.0)
Requirement already satisfied: duet~=0.2.8 in /usr/local/lib/python3.10/dist-packages (from cirq-core~=1.0->openfermion>=0.5->openfermionpyscf==0.5) (0.2.9)
Requirement already satisfied: urllib3<3,>=1.21.1 in /usr/local/lib/python3.10/dist-packages (from requests>=2.18->openfermion>=0.5->openfermionpyscf==0.5) (2.2.1)
Requirement already satisfied: idna<4,>=2.5 in /usr/local/lib/python3.10/dist-packages (from requests>=2.18->openfermion>=0.5->openfermionpyscf==0.5) (3.7)
Requirement already satisfied: certifi>=2017.4.17 in /usr/local/lib/python3.10/dist-packages (from requests>=2.18->openfermion>=0.5->openfermionpyscf==0.5) (2024.2.2)
Requirement already satisfied: charset-normalizer<4,>=2 in /usr/local/lib/python3.10/dist-packages (from requests>=2.18->openfermion>=0.5->openfermionpyscf==0.5) (3.3.2)
Requirement already satisfied: mpmath>=0.19 in /usr/local/lib/python3.10/dist-packages (from sympy->openfermion>=0.5->openfermionpyscf==0.5) (1.3.0)
Requirement already satisfied: tzdata>=2022.7 in /usr/local/lib/python3.10/dist-packages (from pandas->cirq-core~=1.0->openfermion>=0.5->openfermionpyscf==0.5) (2024.1)
Requirement already satisfied: pytz>=2020.1 in /usr/local/lib/python3.10/dist-packages (from pandas->cirq-core~=1.0->openfermion>=0.5->openfermionpyscf==0.5) (2024.1)
Installing collected packages: scipy
Attempting uninstall: scipy
Found existing installation: scipy 1.10.1
Uninstalling scipy-1.10.1:
Successfully uninstalled scipy-1.10.1
Successfully installed scipy-1.13.0
WARNING: Running pip as the 'root' user can result in broken permissions and conflicting behaviour with the system package manager. It is recommended to use a virtual environment instead: https://pip.pypa.io/warnings/venv
[2]:
import cudaq
import matplotlib.pyplot as plt
from scipy.optimize import minimize
import numpy as np
# Single precision
cudaq.set_target("nvidia")
# Double precision
#cudaq.set_target("nvidia-fp64")
---------------------------------------------------------------------------
RuntimeError Traceback (most recent call last)
Cell In[2], line 7
4 import numpy as np
6 # Single precision
----> 7 cudaq.set_target("nvidia")
8 # Double precision
9 #cudaq.set_target("nvidia-fp64")
RuntimeError: [custatevec] %CUDA driver version is insufficient for CUDA runtime version in CuStateVecCircuitSimulator (line 334)
The problem of interest here is a chain of hydrogen atoms seperated along the z-axis at a fixed interval called the bond distance.
The interatomic electrostatic forces due to the electrons and protons and the shielding by the neutrons creates a chemical system whose energy can be minimised to find a stable configuration.
Let us first begin by defining the molecule and other metadata about the problem.
[3]:
# Number of hydrogen atoms.
hydrogen_count = 2
# Distance between the atoms in Angstroms.
bond_distance = 0.7474
# Define a linear chain of Hydrogen atoms
geometry = [('H', (0, 0, i * bond_distance)) for i in range(hydrogen_count)]
molecule, data = cudaq.chemistry.create_molecular_hamiltonian(
geometry, 'sto-3g', 1, 0)
electron_count = data.n_electrons
qubit_count = 2 * data.n_orbitals
We now generate a Unitary Coupled-Cluster Singles and Doubles (UCCSD) ansatz from the template provided by CUDA-Q.
[4]:
@cudaq.kernel
def kernel(thetas: list[float]):
qubits = cudaq.qvector(qubit_count)
for i in range(electron_count):
x(qubits[i])
cudaq.kernels.uccsd(qubits, thetas, electron_count, qubit_count)
parameter_count = cudaq.kernels.uccsd_num_parameters(electron_count,
qubit_count)
Using CUDA-Q Optimizers¶
We use the builtin optimizers within CUDA-Q for the minimization procedure.
[5]:
optimizer = cudaq.optimizers.COBYLA()
energy, parameters = cudaq.vqe(kernel,
molecule,
optimizer,
parameter_count=parameter_count)
print(energy)
-1.1371756649403284
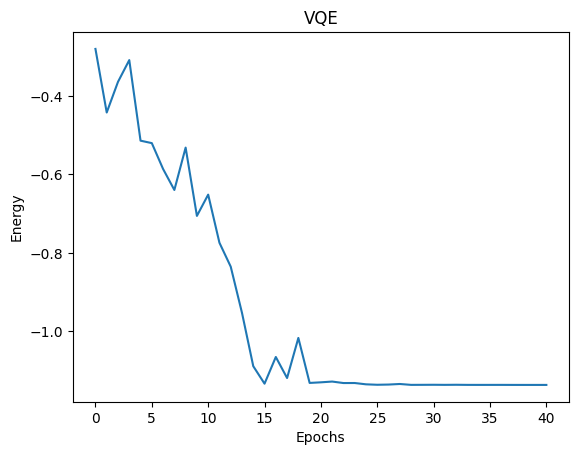
Integration with Third-Party Optimizers¶
We can also integrate popular libraries like scipy with CUDA-Q.
[6]:
# Define a function to minimize
def cost(theta):
exp_val = cudaq.observe(kernel, molecule, theta).expectation()
return exp_val
exp_vals = []
def callback(xk):
exp_vals.append(cost(xk))
# Initial variational parameters.
np.random.seed(42)
x0 = np.random.normal(0, np.pi, parameter_count)
# Use the scipy optimizer to minimize the function of interest
result = minimize(cost,
x0,
method='COBYLA',
callback=callback,
options={'maxiter': 40})
plt.plot(exp_vals)
plt.xlabel('Epochs')
plt.ylabel('Energy')
plt.title('VQE')
plt.show()