QM/MM simulation: VQE within a Polarizable Embedded Framework.¶
In this tutorial, we will discuss a hybrid quantum-classical algorithm called PE-VQE-SCF introduced in this paper. The PE-VQE-scf approch combines:
Variational Quantum Eigensolver (VQE) – a quantum algorithm for finding ground-state energies of molecules.
Self-Consistent Field (SCF) – a classical iterative method used in quantum chemistry.
Polarizable Embedding (PE) – a technique to model the effect of an environment (like a solvent or protein) on a quantum system.
The goal is to simulate chemical systems on quantum computers (or simulator) by embedding them in a polarizable environment. In this tutorial, we will use CUDA-Q to implement the QM/MM framework and the CUDA-Q GPU accelerated simulator for running the simulation.
Key concepts:¶
1- Variational Quantum Eigensolver (VQE) - A quantum algorithm that estimates the ground state energy of a molecule. - It uses a parameterized quantum circuit (ansatz) and a classical optimizer to minimize the energy expectation value. - In this tutorial, we employ VQE with UCCSD ansatz for the quantum part. However, user should be able to replace the VQE and the ansatz with other quantum algorithm.
2- Self-Consistent Field (SCF) - A method where the solution (e.g., molecular orbitals) is updated iteratively until convergence. - In this context, SCF is used to update the embedding potential based on the quantum system’s density.
3- Polarizable Embedding (PE) - Models the environment as a set of polarizable sites that respond to the quantum system’s electric field. - The environment affects the quantum system, and vice versa, requiring mutual polarization.
PE-VQE-SCF Algorithm Steps¶
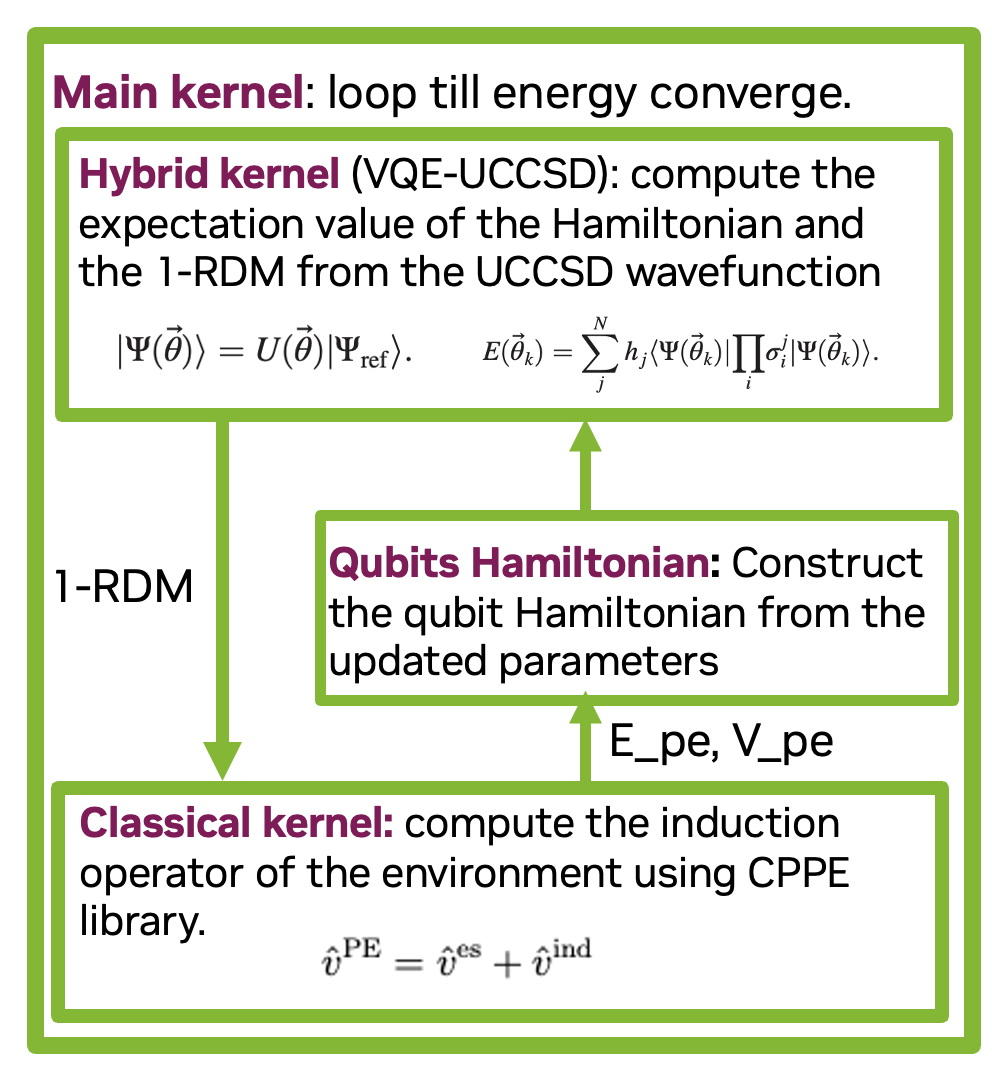
Step 1: Initialize (Classical pre-processing)¶
Define the quantum region (e.g., a molecule) and the classical environment (e.g., solvent).
Set up the PE parameters (multipoles, polarizabilities).
Step 2: Build the Hamiltonian¶
Construct the PE-augmented Hamiltonian that includes the molecular Hamiltonian and the interaction with the polarizable environment.
Step 3: Run VQE¶
Use a quantum computer (or simulator) to estimate the ground state energy of the system using VQE.
The quantum circuit prepares a trial wavefunction, and the classical optimizer adjusts parameters to minimize energy.
Step 4: Update Environment¶
Compute the electronic density from the VQE result.
Update the induced dipoles in the environment based on this density.
Step 5: Self-Consistency Loop¶
Repeat Steps 2–4 until the energy and density converge (i.e., SCF convergence).
Requirments:¶
[ ]:
# You might need to install (apt-get update & apt-get install build-essential -y
# apt-get install python3-dev) before installing cppe.
%pip install cppe
%pip install pyscf
[1]:
from pyscf import gto, scf, solvent
import numpy as np
from functools import reduce
import cudaq
cudaq.set_target("nvidia", option="fp64")
Example 1: LiH with 2 water molecules.¶
Initialize and build the hamiltonian.¶
[6]:
from qchem.classical_pyscf import get_mol_pe_hamiltonian
geometry = 'Li 0.3925 0.0 0.0; H -1.1774 0.0 0.0'
water_pot= "qchem/4NP_in_water.pot"
molecular_data=get_mol_pe_hamiltonian(xyz=geometry, potfile=water_pot, spin=0, charge=0, basis='sto3g', ccsd=True, verbose=True)
obi_mol = molecular_data[0]
tbi_mol = molecular_data[1]
e_nn = molecular_data[2]
obi_pe = molecular_data[3]
nelectrons = molecular_data[4]
norbitals = molecular_data[5]
qubits_num = 2 * norbitals
overwrite output file: Li 0-pyscf.log
[Pyscf] Total number of electrons = 4
[Pyscf] Total number of orbitals = 6
[Pyscf] Total HF energy with solvent: -7.862708330327407
[Pyscf] Polarizable embedding energy from HF: -1.1127389358871828e-05
[Pyscf] Total CCSD energy with solvent: -7.882680504632604
[Pyscf] Polarizable embedding energy from CCSD: -1.0898134916648752e-05
Running solver for induced moments.
0 --- Norm: 0.000095431562
1 --- Norm: 0.000000927586
--- Turning on DIIS. ---
2 --- Norm: 0.000000010648
3 --- Norm: 0.000000000133
[3]:
from qchem.hamiltonian import jordan_wigner_fermion, jordan_wigner_pe
spin_mol_ham=jordan_wigner_fermion(obi_mol, tbi_mol, e_nn, tolerance = 1e-12)
spin_pe_ham=jordan_wigner_pe(obi_pe, tolerance = 1e-12)
[4]:
mol = gto.M(
atom = geometry,
spin = 0,
charge = 0,
basis = 'sto3g'
)
mf = scf.RHF(mol)
mf_pe = solvent.PE(mf, water_pot).run()
converged SCF energy = -7.86270833032741
VQE, update environment, and scf loop.¶
Note: for more accurate result, user needs to change conv_tol to much smaller value, e.g., 1e-7. In addition, the tolerance for the classical optimizer vqe_tol should be modified to smaller value, e.g., 1e-7.
[5]:
from qchem.uccsd import uccsd_parameter_size
from qchem.uccsd_vqe import uccsd_circuit_vqe
from qchem.particle_operator import one_particle_op
from qchem.PEoperator import pe_operator
from qchem.uccsd_init_param import get_parameters
e_last = 0.0
conv_tol = 1e-2
dE = 1.0
cycle = 1
vqe_tol = 1e-4
# Get the number of parameters
singles, doubles, total = uccsd_parameter_size(nelectrons, qubits_num, spin_mult = 0)
print(f"Number of parameters: {singles} singles, {doubles} doubles, {total} total")
# Get the initial parameters for the UCCSD circuit
theta = get_parameters(xyz=geometry, spin=0, charge=0, basis='sto3g', ccsd=True, without_solvent=False, potfile=water_pot)
#theta = np.zeros(total, dtype=np.float64)
while dE > conv_tol:
#for i in range (2):
spin_ham = spin_mol_ham + spin_pe_ham
vqe_result = uccsd_circuit_vqe(spin_mult = 0, only_singles = False, only_doubles = False, qubits_num = qubits_num,
electron_count = nelectrons, optimize = True, theta = theta,
hamiltonian = spin_ham, method = 'L-BFGS-B', vqe_tol = vqe_tol, verbose = False)
print(f"VQE Energy: {vqe_result[0]}")
#print(f"VQE parameters: {vqe_result[1]}")
print(f"VQE converged: {vqe_result[2]}")
theta = vqe_result[1]
if vqe_result[2]:
# Compute rdm in atomic orbital.
dm_a = np.zeros((qubits_num // 2, qubits_num // 2))
dm_b = np.zeros((qubits_num // 2, qubits_num // 2))
n_spatial_orbitals = qubits_num // 2
n_occupied = nelectrons // 2
n_virtual = n_spatial_orbitals - n_occupied
alpha_indices = [i * 2 for i in range(n_occupied)]
alpha_indices += [i * 2 + nelectrons for i in range(n_virtual)]
beta_indices = [i * 2 + 1 for i in range(n_occupied)]
beta_indices += [i * 2 + 1 + nelectrons for i in range(n_virtual)]
for i in range(len(alpha_indices)):
p = alpha_indices[i]
for j in range(len(alpha_indices)):
q = alpha_indices[j]
qubit_op_dm = one_particle_op(p, q)
result = uccsd_circuit_vqe(spin_mult = 0, only_singles = False, only_doubles = False, qubits_num = qubits_num,
electron_count = nelectrons, optimize = False, theta = theta,
hamiltonian = qubit_op_dm, verbose = False)
dm_a[i, j] = result[0]
for i in range(len(beta_indices)):
p = beta_indices[i]
for j in range(len(beta_indices)):
q = beta_indices[j]
qubit_op_dm = one_particle_op(p, q)
result = uccsd_circuit_vqe(spin_mult = 0, only_singles = False, only_doubles = False, qubits_num = qubits_num,
electron_count = nelectrons, optimize = False, theta = theta,
hamiltonian = qubit_op_dm, verbose = False)
dm_b[i, j] = result[0]
dm_ao = dm_a + dm_b
dm_ao = reduce(np.dot, (mf_pe.mo_coeff, dm_ao, mf_pe.mo_coeff.T))
spin_pe_ham, E_pe, V_pe = pe_operator(dm_ao, mol, mf_pe.mo_coeff, water_pot, tolerance = 1e-12)
result = uccsd_circuit_vqe(spin_mult = 0, only_singles = False, only_doubles = False, qubits_num = qubits_num,
electron_count = nelectrons, optimize = False, theta = theta,
hamiltonian = spin_pe_ham, verbose = False)
edup = result[0]
# Check energy are the same
ovlp = mol.intor_symmetric('int1e_ovlp')
dm = np.einsum('pi,pq,qj->ij', ovlp, dm_ao, ovlp)
dm_mo = reduce(np.dot, (mf_pe.mo_coeff.T, dm, mf_pe.mo_coeff))
edup_np = np.einsum('ij,ji->', V_pe, dm_mo)
print('Energy from observe call= {:15g} & Energy from np einsum= {:15g}' .format(edup, edup_np), flush=True)
e_current = vqe_result[0] - edup + E_pe
dE = np.abs(e_current - e_last)
e_last = e_current
print('Cycle {:d} E_tot = {:.15g} dE = {:g}' .format(cycle, e_last, dE), flush=True)
print('Polarizable embedding energy: ', E_pe, flush=True)
print('Expectation value of polarizable operator: ', edup, flush=True)
print('\n')
cycle+=1
else:
#raise ValueError("Unsuccessful optimization.")
print("Unsuccessful optimization. Restart the optimization.", flush=True)
pass
# Compute the RDM in molecular orbitals and natural orbital occupation number.
ovlp = mol.intor_symmetric('int1e_ovlp')
dm = np.einsum('pi,pq,qj->ij', ovlp, dm_ao, ovlp)
dm_mo = reduce(np.dot, (mf_pe.mo_coeff.T, dm, mf_pe.mo_coeff))
noon, U = np.linalg.eigh(dm_mo)
noon = np.flip(noon)
print('\n')
print('Final result: ')
print('Polarizable embedding energy: ', E_pe)
print('Expectation value of polarizable operator: ', edup, flush=True)
print('PE-VQE-UCCSD energy (L-BFGS-B)= ', e_last)
print('Natural orbitals occupation number: ', noon)
Number of parameters: 16 singles, 76 doubles, 92 total
overwrite output file: Li 0-pyscf.log
Optimizer exited successfully: True
VQE Energy: -7.867161909761761
VQE converged: True
Running solver for induced moments.
0 --- Norm: 0.000095457621
1 --- Norm: 0.000000860362
--- Turning on DIIS. ---
2 --- Norm: 0.000000004312
Energy from observe call= 0.000762169 & Energy from np einsum= 0.000762197
Cycle 1 E_tot = -7.86793486027634 dE = 7.86793
Polarizable embedding energy: -1.0781673490428836e-05
Expectation value of polarizable operator: 0.0007621688410912728
Optimizer exited successfully: True
VQE Energy: -7.8692404296251395
VQE converged: True
Running solver for induced moments.
0 --- Norm: 0.000095564807
1 --- Norm: 0.000000887381
--- Turning on DIIS. ---
2 --- Norm: 0.000000007916
Energy from observe call= 0.000762218 & Energy from np einsum= 0.000762229
Cycle 2 E_tot = -7.87001339604889 dE = 0.00207854
Polarizable embedding energy: -1.0748496723151968e-05
Expectation value of polarizable operator: 0.0007622179270272233
Final result:
Polarizable embedding energy: -1.0748496723151968e-05
Expectation value of polarizable operator: 0.0007622179270272233
PE-VQE-UCCSD energy (L-BFGS-B)= -7.87001339604889
Natural orbitals occupation number: [1.99899690e+00 1.91601348e+00 6.71542420e-02 1.60555115e-02
1.15390205e-03 6.30790416e-04]
Example 2: NH3 with 46 water molecule using active space.¶
[2]:
from qchem.classical_pyscf import get_mol_pe_hamiltonian
from qchem.hamiltonian import jordan_wigner_fermion, jordan_wigner_pe
geometry_NH3 = 'qchem/NH3.xyz'
water_pot = 'qchem/46_water.pot'
molecular_data=get_mol_pe_hamiltonian(xyz=geometry_NH3, potfile=water_pot, spin=0, charge=0, basis='631g', \
nele_cas=6, norb_cas=6, ccsd=True, verbose=True)
obi_mol = molecular_data[0]
tbi_mol = molecular_data[1]
e_core = molecular_data[2]
obi_pe =molecular_data[3]
nelectrons = molecular_data[4]
norbitals = molecular_data[5]
qubits_num = 2 * norbitals
spin_mol_ham = jordan_wigner_fermion(obi_mol, tbi_mol, e_core)
spin_pe_ham = jordan_wigner_pe(obi_pe)
overwrite output file: qchem/NH3-pyscf.log
[Pyscf] Total number of electrons = 10
[Pyscf] Total number of orbitals = 15
[Pyscf] Total HF energy with solvent: -56.460544146152934
[Pyscf] Polarizable embedding energy from HF: -0.2987297397877397
[pyscf] CCSD energy of the active space (using molecular orbitals) with solvent= -56.47023410365107
[Pyscf] Polarizable embedding energy from CCSD: -0.2986368707105786
Running solver for induced moments.
0 --- Norm: 0.572427861637
1 --- Norm: 0.194328219249
--- Turning on DIIS. ---
2 --- Norm: 0.063478479823
3 --- Norm: 0.007602903988
4 --- Norm: 0.002247104251
5 --- Norm: 0.000460072303
6 --- Norm: 0.000114086983
7 --- Norm: 0.000023144988
8 --- Norm: 0.000005162818
9 --- Norm: 0.000001149278
10 --- Norm: 0.000000248690
11 --- Norm: 0.000000069750
12 --- Norm: 0.000000018086
13 --- Norm: 0.000000001401
[3]:
from pyscf import gto, scf, mcscf, solvent
mol = gto.M(
atom = geometry_NH3,
spin = 0,
charge = 0,
basis = '631g'
)
mf_pe = scf.RHF(mol)
mf_pe = solvent.PE(mf_pe, water_pot).run()
mc = mcscf.CASCI(mf_pe, norbitals, nelectrons)
converged SCF energy = -56.460544146164
Note: for more accurate result, user needs to change conv_tol to much smaller value, e.g., 1e-7. In addition, the tolerance for the classical optimizer vqe_tol should be modified to smaller value, e.g., 1e-7.
[4]:
import numpy as np
from functools import reduce
from qchem.uccsd import uccsd_parameter_size
from qchem.uccsd_vqe import uccsd_circuit_vqe
from qchem.particle_operator import one_particle_op
from qchem.PEoperator import pe_operator_as
from qchem.uccsd_init_param import get_parameters
# Get the number of parameters
singles, doubles, total = uccsd_parameter_size(nelectrons, qubits_num, spin_mult = 0)
print(f"Number of parameters: {singles} singles, {doubles} doubles, {total} total")
#theta = get_parameters(xyz = geometry_NH3, spin = 0,charge = 0, basis = '631g', nele_cas = 6, norb_cas = 6, \
# ccsd = True, without_solvent = False, potfile = water_pot)
theta = np.zeros(total, dtype=np.float64)
e_last = 0.0
conv_tol = 1e-2
dE = 1.0
cycle = 1
vqe_tol = 1e-3
while dE > conv_tol:
#for i in range (1):
spin_ham = spin_mol_ham + spin_pe_ham
vqe_result = uccsd_circuit_vqe(spin_mult = 0, only_singles = False, only_doubles = False, qubits_num = qubits_num,
electron_count = nelectrons, optimize = True, theta = theta,
hamiltonian = spin_ham, method = 'L-BFGS-B', vqe_tol = vqe_tol, verbose = False)
print(f"VQE Energy: {vqe_result[0]}")
#print(f"VQE parameters: {vqe_result[1]}")
print(f"VQE converged: {vqe_result[2]}")
theta = vqe_result[1]
if vqe_result[2]:
dm_a = np.zeros((qubits_num // 2, qubits_num // 2))
dm_b = np.zeros((qubits_num // 2, qubits_num // 2))
dm = np.zeros((qubits_num, qubits_num))
n_spatial_orbitals = qubits_num // 2
n_occupied = nelectrons // 2
n_virtual = n_spatial_orbitals - n_occupied
alpha_indices = [i * 2 for i in range(n_occupied)]
alpha_indices += [i * 2 + nelectrons for i in range(n_virtual)]
beta_indices = [i * 2 + 1 for i in range(n_occupied)]
beta_indices += [i * 2 + 1 + nelectrons for i in range(n_virtual)]
for i in range(len(alpha_indices)):
p = alpha_indices[i]
for j in range(len(alpha_indices)):
q = alpha_indices[j]
qubit_op_dm = one_particle_op(p, q)
result = uccsd_circuit_vqe(spin_mult = 0, only_singles = False, only_doubles = False, qubits_num = qubits_num,
electron_count = nelectrons, optimize = False, theta = theta,
hamiltonian = qubit_op_dm, verbose = False)
dm_a[i, j] = result[0]
for i in range(len(beta_indices)):
p = beta_indices[i]
for j in range(len(beta_indices)):
q = beta_indices[j]
qubit_op_dm = one_particle_op(p, q)
result = uccsd_circuit_vqe(spin_mult = 0, only_singles = False, only_doubles = False, qubits_num = qubits_num,
electron_count = nelectrons, optimize = False, theta = theta,
hamiltonian = qubit_op_dm, verbose = False)
dm_b[i, j] = result[0]
mocore = mf_pe.mo_coeff[:, :mc.ncore]
dm_core = np.dot(mocore, mocore.conj().T) * 2
casdm = dm_a + dm_b
mocas = mf_pe.mo_coeff[:, mc.ncore:mc.ncore + mc.ncas]
# RDM in atomic orbitals
dm = dm_core + reduce(np.dot, (mocas, casdm, mocas.conj().T))
spin_pe_hamiltonian, E_pe, V_pe, V_pe_cas= pe_operator_as(dm,mol,mf_pe.mo_coeff,water_pot,mc)
result = uccsd_circuit_vqe(spin_mult = 0, only_singles = False, only_doubles = False, qubits_num = qubits_num,
electron_count = nelectrons, optimize = False, theta = theta,
hamiltonian = spin_pe_ham, verbose = False)
edup = result[0]
e_current = vqe_result[0] - edup + E_pe
dE = np.abs(e_current - e_last)
e_last = e_current
print('Cycle {:d} E_tot = {:.15g} dE = {:g}' .format(cycle, e_last, dE), flush=True)
print('Polarizable embedding energy: ', E_pe, flush=True)
print('Expectation value of polarizable operator: ', edup, flush=True)
print('\n')
cycle+=1
else:
print("Unsuccessful optimization. Restart the optimization.", flush=True)
pass
# convert dm from ao to mo (D_MO= C.T S D_AO S C ) and compute natural orbital occupation number.
ovlp = mol.intor_symmetric('int1e_ovlp')
dm = np.einsum('pi,pq,qj->ij', ovlp, dm, ovlp)
dm_mo = reduce(np.dot, (mf_pe.mo_coeff.T, dm, mf_pe.mo_coeff))
noon, U = np.linalg.eigh(dm_mo)
noon = np.flip(noon)
print('\n')
print('Final result: ')
print('Polarizable embedding energy: ', E_pe)
print('Expectation value of polarizable operator: ', edup, flush=True)
print('PE-VQE-UCCSD energy (L-BFGS-B)= ', e_last)
print('Natural orbitals occupation number: ', noon)
Number of parameters: 18 singles, 99 doubles, 117 total
Optimizer exited successfully: True
VQE Energy: -56.07492192212563
VQE converged: True
Running solver for induced moments.
0 --- Norm: 0.560904998895
1 --- Norm: 0.195464440561
--- Turning on DIIS. ---
2 --- Norm: 0.080909047471
3 --- Norm: 0.014193006098
4 --- Norm: 0.004526458635
5 --- Norm: 0.001256746907
6 --- Norm: 0.000476278998
7 --- Norm: 0.000120262763
8 --- Norm: 0.000033369845
9 --- Norm: 0.000007548513
10 --- Norm: 0.000001932468
11 --- Norm: 0.000000513677
12 --- Norm: 0.000000251862
13 --- Norm: 0.000000062997
14 --- Norm: 0.000000031247
15 --- Norm: 0.000000008129
Cycle 1 E_tot = -56.4704454234038 dE = 56.4704
Polarizable embedding energy: -0.29871334470788147
Expectation value of polarizable operator: 0.09681015657024516
Optimizer exited successfully: True
VQE Energy: -56.074941291297016
VQE converged: True
Running solver for induced moments.
0 --- Norm: 0.578438887906
1 --- Norm: 0.186313099889
--- Turning on DIIS. ---
2 --- Norm: 0.068418421402
3 --- Norm: 0.012730202853
4 --- Norm: 0.004094551612
5 --- Norm: 0.000999816754
6 --- Norm: 0.000319250864
7 --- Norm: 0.000077598802
8 --- Norm: 0.000023880904
9 --- Norm: 0.000008037391
10 --- Norm: 0.000001859332
11 --- Norm: 0.000000532803
12 --- Norm: 0.000000133206
13 --- Norm: 0.000000032446
14 --- Norm: 0.000000009283
Cycle 2 E_tot = -56.4704648912704 dE = 1.94679e-05
Polarizable embedding energy: -0.2987133198747535
Expectation value of polarizable operator: 0.09681028009861878
Final result:
Polarizable embedding energy: -0.2987133198747535
Expectation value of polarizable operator: 0.09681028009861878
PE-VQE-UCCSD energy (L-BFGS-B)= -56.47046489127038
Natural orbitals occupation number: [ 2.00000000e+00 2.00000000e+00 1.99935887e+00 1.99541659e+00
1.99508319e+00 4.74450015e-03 4.36742635e-03 1.08093504e-03
4.89965694e-15 1.85085120e-15 1.30851294e-15 5.07383839e-16
-1.05732651e-15 -1.59225347e-15 -6.12772938e-15]
[5]:
print(cudaq.__version__)
CUDA-Q Version cu12-latest (https://github.com/NVIDIA/cuda-quantum 615d91910310605c83ea59f6afe6e7ae6dfecd28)